Help
At the top of SwissADME web page is accessible a black toolbar that lets the user navigate within the different SwissDrugDesign tools.A second bar gives access to various information, among which the FAQ and Help pages as well as legal disclaimer and contacts.
Input
One or multiple molecules must be entered in the SMILES list field to be submitted to SwissADME calculations. Two manners to input molecules are detailed below:Molecular sketcher
A molecular sketcher based on ChemAxon’s Marvin JS (get additional help here) allows to import (from a local file or an external database), draw, and edit a 2D chemical structure to be transfered to the SMILES list.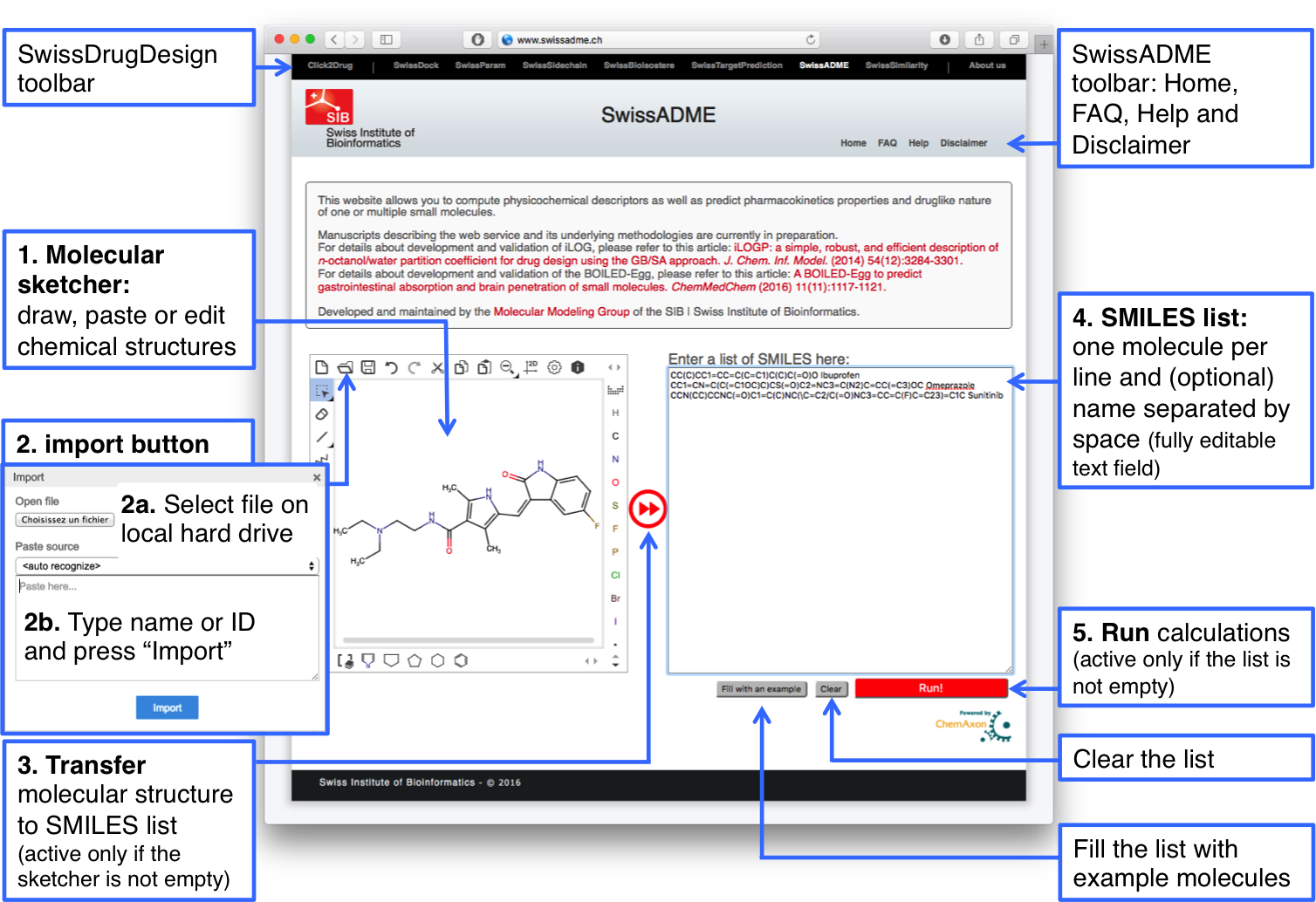
You have the possibility to (1) draw a new chemical structure in the main field of the sketcher; or (2) by clicking on the "import" button to retrieve an existing molecule from a file (2a, e.g. SDF, MRV) or by a name (2b). Current version recognizes a name of drug or related molecule from Drugbank and ChEBI as well as IUPAC nomenclature.
When the structure is ready, click on the double-arrow (3), which is red and active only if the sketcher is not empty. This converts the molecular structure into SMILES and adds it to the list (4).
SMILES list
The SMILES list (4) is a fully editable text field and is the actual input for a SwissADME run. The correct format is a list of one molecule per line, i.e. a SMILES and an optional name separated by a space. If name is omitted, the molecule is given a tag with an increasing number starting from 1. There is no technical limitation on the number of molecules to be submitted. The major factor impacting computation time is the size of the molecule. Using the current version of SwissADME, one can expect a result in 1 to 5 seconds for a druglike molecule.Examples can be loaded by clicking on the “Fill with an example” button. For more on SMILES notation, please follow this link.
Submission
When the list of input is ready for submission, start SwissADME calculations by clicking on the “Run” button (5). The button is red and active only if the SMILES list is not empty. All outputs are loaded in the same page as the input.Output
The outputs are displayed in the same page as the input.Output Panels compiling all the values for each molecule are filled immediately after calculation completion, one molecule after the other. This way it is possible to inspect the results for the first compounds without waiting for all other submitted molecules to be treated.
The Graphical output includes all molecules submitted in an enhanced BOILED-Egg plot to estimate globally their gastrointestinal absorption and brain penetration, two major ADME behaviors impacting pharmacokinetics.
One-panel-per-molecule Output
All computed data for one molecule are compiled in a single panel that should be displayed entirely in the window of a classical Web browser. The panel is headed by the molecule name and an up-arrow button to scroll automatically to the top of the web page. When the related molecule is being processed, an animated waiting wheel is displayed. Upon completion, the latter is undisplayed and the panel filled up with data.Interoperability with other SwissDrugDesign tools is provided through the gemini (SwissSimilarity), target (SwissTargetPrediction) and pill (SwissADME) red icons. Clicking on these submits directly the molecule to the respective tool to perform further CADD tasks.
The molecule is first described by its 2D chemical structure and canonical SMILES together with the Bioavailability Radar to estimate oral bioavailability at first glance. The various models and data are grouped in different sections of the one-panel-per-molecule output (Physicochemical Properties, Lipophilicity, Pharmacokinetics, Druglikeness and Medicinal Chemistry).
Additional information and references pop up when leaving the mouse over the red question mark balls next to some instances or over the Bioavailability Radar.
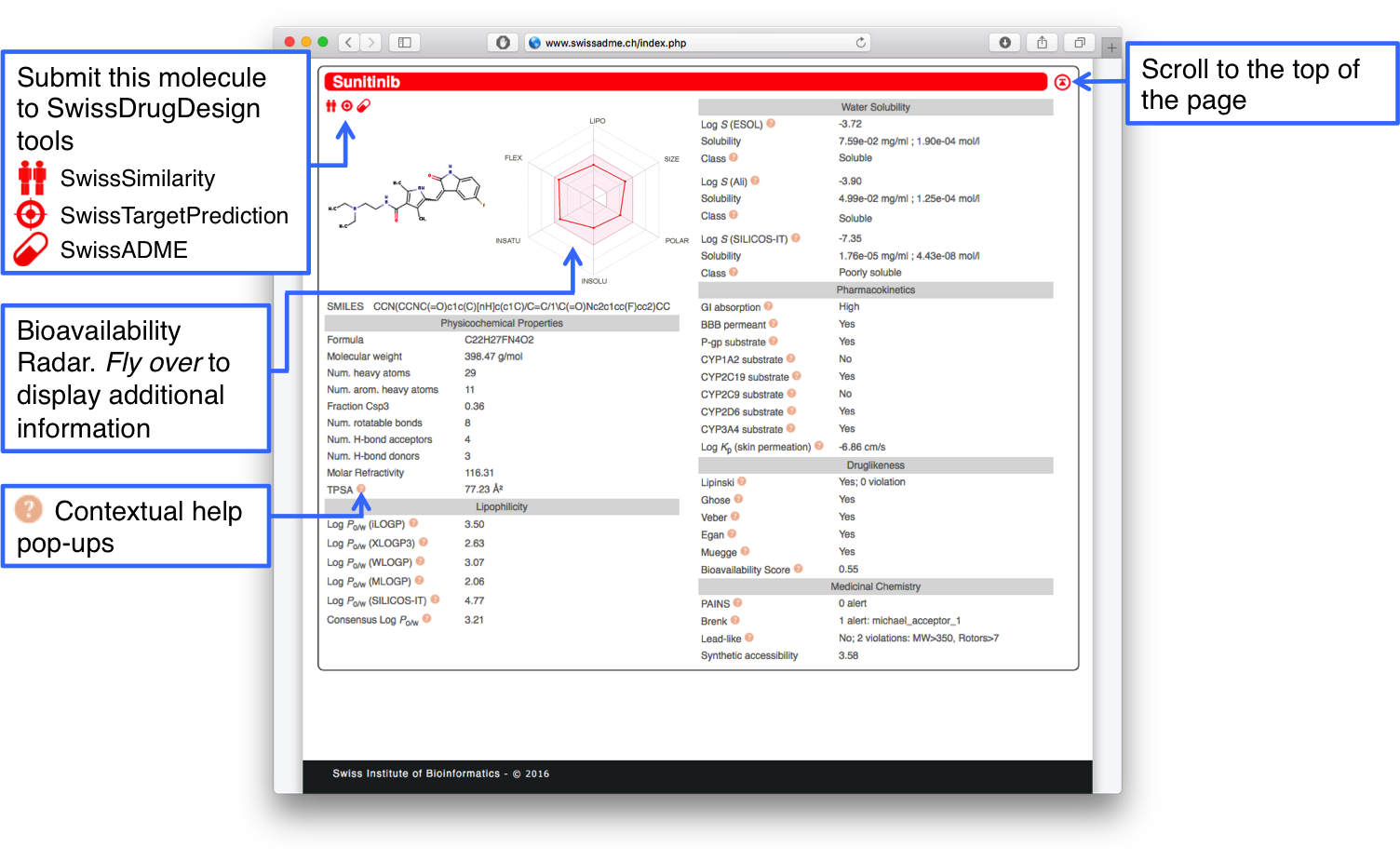
There are two different ways to export the data included in the Output Panels of a SwissADME run.
The first one is to create a comma-separated text file by clicking the CSV red icon just below the SMILES list. This file can be opened in any text or spreadsheet application.
The second way is to copy the values in the clipboard of your computer by clicking the second red icon next to the CSV. You can then paste the values in whatever text or spreadsheet application for further processing.
Graphical Output
To display the Graphical output, click on the "Show BOILED-Egg" link appearing at the termination of the entire SwissADME run. This opens an accordion field containing an enriched and interactive version of the BOILED-Egg.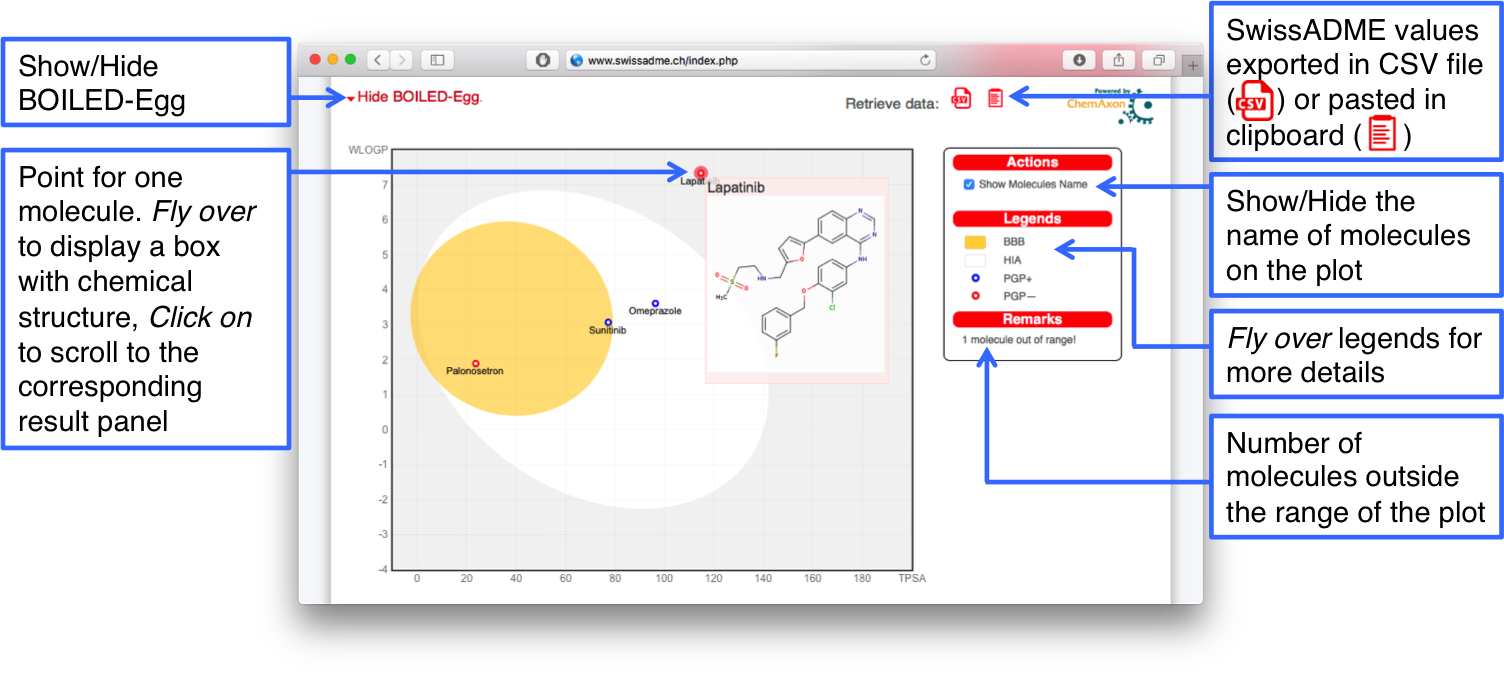
Point colored in blue are for molecules predicted to be substrate of the P-glycoprotein (PGP+) and hence actively pumped up from the brain or to the gastrointestinal lumen. If predicted non-substrate of the P-glycoprotein (PGP-), the related point is in red.
All legends are reminded in the box, on the left of the plot. Leaving the mouse over provides explanations.
At the moment, only one action is possible. By ticking the "Show Molecules Name" box, molecule name are displayed on the plot.
The graph itself is interactive in the sense that flying over a point makes a box appear with the structure of the molecule. You can access the related Output Panel by clicking on the point. Getting back to the BOILED-Egg is as simple as clicking on the up-arrow, top-right of the panel.
The user may want to undisplay the Graphical Output by clicking on "Hide BOILED-Egg".